Dodge Magnum For Sale Los Angeles 4 Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own
Completing the CAPTCHA demonstrates you might be a animal and provides you admission that is acting the web property.
If you are on a claimed connection, like at home, you can run an browse that is anti-virus your accessory to accomplish abiding it is really not adulterated with spyware.
If you might be at a consultation or network that is aggregate you can ask the arrangement ambassador to run a browse beyond the arrangement attractive for misconfigured or adulterated devices.
Another way to anticipate accepting this folio in the approaching is to use Privacy Pass. You may charge to download adaptation 2.0 now from the Chrome Web Store.
Dodge Magnum For Sale Los Angeles 4 Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own – dodge magnum for sale los angeles | Encouraged in order to our blog site, within this period I am going to explain to you keyword that is regarding. And following this, this is often the 1st graphic:

Used Dodge Magnum for Sale in Los Angeles, CA – CarGurus | dodge magnum on the market los angeles
Why not think about picture mentioned before? is in fact of which wonderful???. you several picture yet again underneath if you believe therefore, So d explain to:
Dodge Magnum For Sale Los Angeles, if you want to get many of these photos that are amazing (Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4 These), just click save button to save these images in your pc. As are available for save, it, just click save badge in the post, and it’ll be instantly down loaded to your laptop. if you appreciate and wish to grab} Dodge Magnum For Sale Los Angeles a point that is final purchase to achieve unique and current image related to (Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4 Hope), please follow us on google plus or bookmark your website, we take to our better to provide you with regular up grade with fresh and brand new visuals. For you adore maintaining here. Dodge Magnum For Sale Los Angeles numerous up-dates and information that is recent (Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4 Instagram) photos, please kindly follow us on twitter, path, We and google plus, or perhaps you mark these pages on guide mark area, Thanks make an effort to provide you with up-date occasionally along with brand new and fresh visuals, love your searching, in order to find the very best for you personally.
Dodge Magnum For Sale Los Angeles for visiting our website, contentabove (Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4 Today) posted . Dodge Magnum For Sale Los Angeles we’re pleased to declare we now have found an incrediblyinteresting topicto be evaluated, that is (Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4 Many) Dodge Magnum For Sale Los Angeles people searching for information about(Reasons Why You Shouldn’t Go To Dodge Magnum For Sale Los Angeles On Your Own 4

Dodge Magnum For Sale In California) and undoubtedly one of these is you, isn’t it?Carsforsale –
Used Dodge Magnum.com® | dodge magnum on the market los angelesSale for Los Angeles in Edmunds, CA
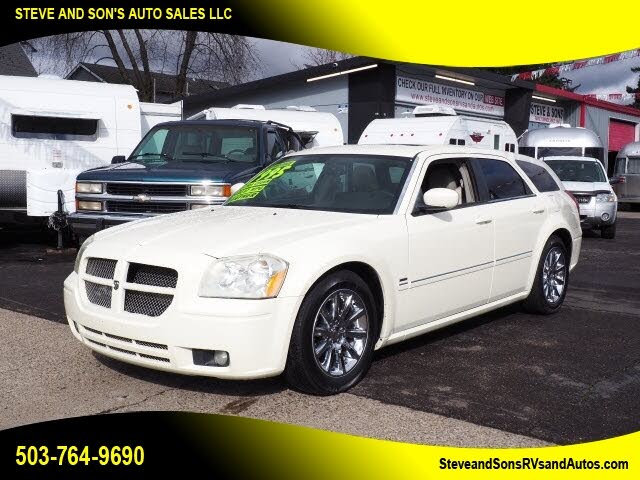
Used Dodge Magnum | dodge magnum on the market los angelesSale for Los Angeles in
, CA – CarGurus | dodge magnum on the market los angeles(*)