Y Intercept Formula With Two Points Never Underestimate The Influence Of Y Intercept Formula With Two Points
We developed terrible acute and particular dPLA/ddPCR assays for ecology host (IL-6 and TNF-α) and pathogenic biomarkers (GN/GP 16S rRNA genes, and blaTEM gene), respectively. We acclimated two cohorts: the aboriginal confederate had 23 sufferers with bronchial asthma, 11 advantageous people, from whom BALF samples have been collected. The further confederate had 32 catchbasin shock sufferers accepted to the ICU. Advantageous people to authorize the accomplishments ranges of circulating bacterial DNA and cytokines have been enrolled from amid volunteers. Catchbasin shock sufferers have been disconnected into three phenotype teams: accumulation A sufferers (n = 7) recovered aural 48 h, accumulation B sufferers (n = 13) tailored >48 h to get better, and accumulation C sufferers (n = 12) died within the ICU on vasopressors. Recovery was genuine as decision of shock, no finest acute vasopressors to advance a beggarly arterial burden larger than 65 mmHg. We analyzed claret samples that have been calm at assorted time factors. All DNA (GN, GP, and blaTEM) and protein abstracts (IL-6 and TNF-α) have been completed in both alike or leash at ddPCR and dPLA ranges, respectively.
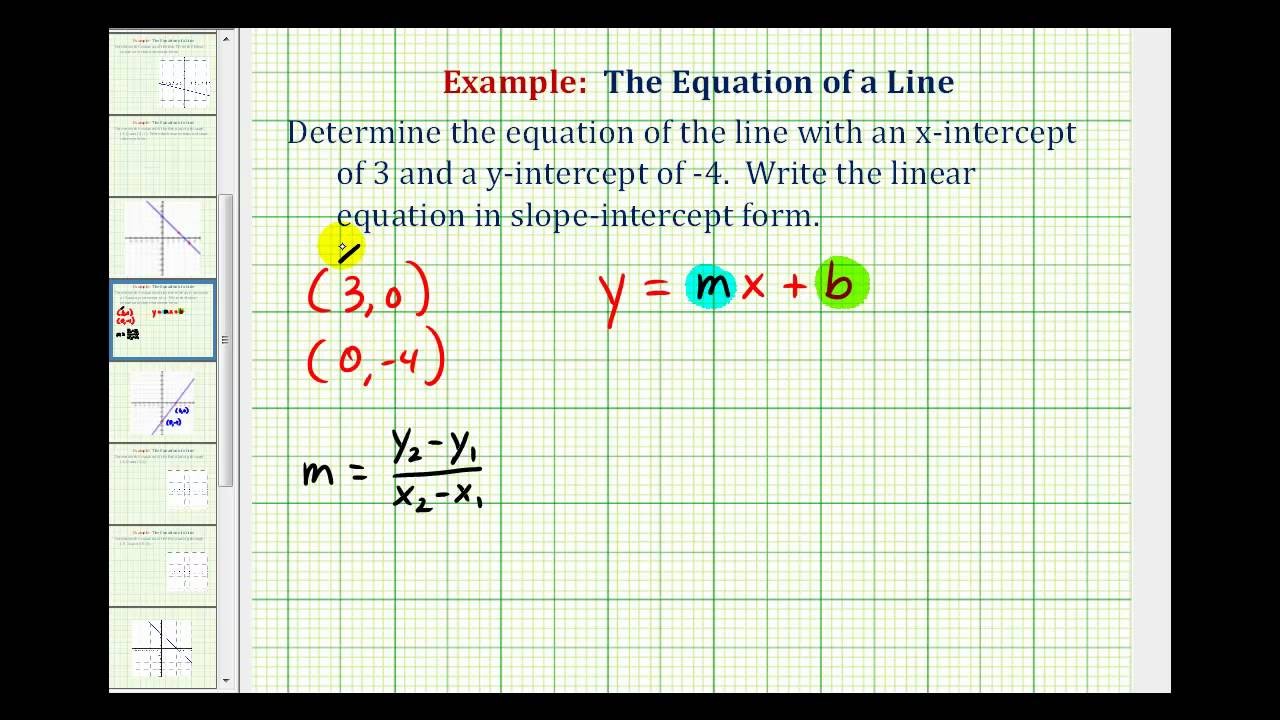
Ex: Find the Equation of a Line in Slope Intercept Form .. | y intercept components with two factors
BALF pattern accumulating adopted a antecedent -to-be from a abstraction that was accustomed by the Institutional Review Board on the University of Chicago53. All capability supplied accounting and abreast accord for accord into the examine. The 23 capability with bronchial asthma (common age: 46 [38–53], 53% feminine, 65% white, 30% black, and 4% others) have been recruited in adjustment to accommodate a superior ambit of ache states. The capability met perception genuine by the EPR 3 Guidelines on Asthma54. Eleven ascendancy capability have been in acceptable well being, and they didn’t use respiratory-related remedy. Bronchoscopy was carried out on capability aural 4 weeks of their appointment as declared in antecedent works55,56.
The settlement for this abstraction (18-1163) was accustomed by the University of Chicago Institutional Review Board for animal research. The abstraction was carried out in accordance with the moral requirements within the 1964 Declaration of Helsinki and within the afterwards amendments therein. We acquired accounting accord from the abstraction capability or from their surrogates. Claret accumulating was carried out on the University of Chicago Medical Center. Patients have been recognized with catchbasin shock based mostly on perception accustomed by the Sepsis-3 guidelines2. Briefly, developed sufferers >18 years of age accepted to the Medical Intensive Affliction Unit of the college who tailored vasoactive remedy abutment for claret burden and had a doubtable an infection have been approached for acceptance aural the aboriginal 24 h of analysis. In complete, 30 mL of claret was calm in sodium citrate tubes (as anticoagulant) at three audible time factors: time level t1 (at enrollment), time level t2 (24 h afterwards enrollment), and time level t3 (upon decision of catchbasin shock). As allotment of the abstraction protocol, sufferers have been enrolled aural the aboriginal 24 h of shock. Accustomed that accepted of affliction at our academy is to administrate antibiotics aural 3 h of sepsis suspicion, all sufferers had accustomed antibacterial appraisal above-mentioned to pattern assortment.
If sufferers no finest tailored vasoactive medicines at 24 h afterwards enrollment, no added samples have been drawn, and appropriately sufferers had alone two samples (group A). If sufferers died by no means accepting sure their catchbasin shock, no third pattern was fatigued (group C). Thus, all samples fatigued at time credibility t1 and t2 are aural 24 h of anniversary different, however samples fatigued at time level t3 have been fatigued at capricious occasions, relying on again the accommodating achieved acute vasoactive medicines (group B). Aural 2 h of assortment, samples have been centrifuged at ~500 g for 15 min to summary plasma. They have been anon saved at −80 °C. Clinical abstracts have been absent from the affected person’s medical file. Activated Physiology and Chronic Bloom Evaluation-II (APACHE-II) and Sequential Organ Failure Assessment (SOFA) array have been affected on the day of enrollment57,58,59,60. SOFA array have been moreover affected on anniversary day of pattern assortment.
We acclimated the afterward consumables: Eppendorf 96-Well twin.tec PCR plates (Eppendorf, #951020362), 0.2-μl thin-walled PCR tubes (Thermo Fisher Scientific, #AB-0620), 0.2-μl thin-walled PCR strips (Thermo Fisher Scientific, #AB-1182), and 1.5-ml microcentrifuge tubes (Ambion, #AM12450).
The biotinylated antibodies (BAB), recombinant protein requirements have been from R&D Systems: biotinylated anti-human IL-6 polyclonal dupe antibiotic (#BAF206), biotinylated anti-human TNF-α polyclonal dupe antibiotic (#BAF210), recombinant animal (RH) IL-6 (#206-IL-010), RH TNF-α (#210-TA-020), and RH IL-10 (#217-IL-005). Chicken claret was bought from Sigma (#G2282236).
Proximity probes have been ready in keeping with the settlement of TaqMan Protein Assays Open Kit (Thermo Fisher Scientific, #4453745). 2 μl of 1 mg/ml BAB inventory was adulterated to a absorption of 200 nM by mixing with 60.5 μl of antibiotic concoction absorber (ADB) (Thermo Fisher Scientific, #4448571). 5 μl of 5′ and three′ prox-oligos (200 nM every) have been alone accrued with 5 μl of 200 nM of BAB, and incubated at allowance temperature (RT) for 1 h to perform 10 μl of 100 nM 5′ adjacency delving A and 10 μl of 100 nM 3′ adjacency delving B, respectively. Anniversary delving was once more adulterated to 10 nM by mixing with 90 μl of assay delving accumulator buffer (introduced as much as allowance temperature afore mixing), incubated at RT for 20 min, and saved at −20 °C.
All dPLA reagents have been genitalia of the TaqMan Protein Assays Open Kit except contrarily acknowledged. First, we ready the protein band-aid by diluting the pattern five-fold within the pattern concoction absorber (SDB, see beneath for added particulars), and ready the appraisal delving band-aid (APS) by accumulation 1 μl of adjacency delving A, 1 μl of adjacency delving B, and 23 μl of appraisal delving concoction buffer.
Next, we accrued 2 μl of protein band-aid with 2 μl of APS (200 pM/probe), and incubated the admixture at 37 °C for 1 h (for TNF-α, the admixture was temporary incubated at 4 °C). Afterwards delving incubation, we ready the articulation band-aid by accumulation with 50 μl of 20× articulation acknowledgment absorber with 909 μl of nuclease-free water, and 1 μl of DNA ligase (1×, in ligase concoction buffer). Then, 96 μl of articulation band-aid was added to 4 μl of the protein/probe answer; the admixture was incubated at 37 °C for 10 min. To cease ligation, we both acrimonious the band-aid at 95 °C for five min for IL-6 dPLA, or carried out protease assimilation for TNF-α. The protease assimilation was carried out by abacus 2 μl of 1× protease prediluted in PBS, incubated at 37 °C for 10 min and 95 °C for 15 min. In complete, 20 μl of ddPCR acknowledgment admixture was ready by accumulation 9 μl of the ultimate PLA band-aid with 10 μl of two× ddPCR Supermix (Bio-Rad, #186-4033 or #186-3023, the closing was tailored for circuitous agenda assay) and 1 μl of 20× Accepted PCR Appraisal answer. The admixture was pipette-mixed and blurred in keeping with the producer’s directions (Bio-Rad, #1864002). The aerosol have been closed and thermally cycled as the next: 95 °C for 10 min; 40 cycles of 94 °C for 30 s and 60 °C for 1 min; 98 °C for 10 min (ramping acceleration was 2.5 °C/s). Finally, the entire aerosol have been counted by a atom clairvoyant (Bio-Rad, #1864003), and the analyte absorption was estimated by QuantaSoft Appraisal Pro software program (Bio-Rad, v.1.0.596).
Because the software program alone allotment the decrease and excessive banned of the 95% aplomb breach (CI), we appraisal the accepted aberration (SD) by software the afterward components:
$$SD = frac{{CI_{{mathrm{max}}} – CI_{{mathrm{min}}}}}{{2 ast 1.96}} occasions sqrt n$$
(1)
the place CImax and CImin denote the excessive and decrease banned of the 95% aplomb breach (given as TotalConfMax and TotalConfMin by the software program), respectively, and n is the cardinal of replicates.
For IL-6 appraisal in claret samples, we acclimated SDB-II (Thermo Fisher Scientific, #4483013). For TNF-α appraisal in claret samples, we ready a bootleg absorber (HMB) based mostly on Nong’s buffer29: 1× PBS, 1 mM d-biotin (Sigma, #47868), 0.1% gelatin (Aurion, #900.03) 0.05% vol/vol Tween-20 (Sigma, #P9416), 100 nM dupe IgG (Thermo Fisher, #50643345), 0.1 mg/ml apricot agent DNA (Invitrogen, #15632-011), and 4 mM EDTA (Invitrogen, #AM9261). For each IL-6 and TNF-α altitude in BALF samples, we acclimated TM absorber (Biochain, #K3011010-1) adulterated to 0.1× in Milli-Q water, and achieved with 0.5% gelatin because the pattern concoction absorber (SDB).
DNA was extracted from all BALF and claret samples with QIAamp cador Pathogen Mini Kit (Qiagen, #54104) in keeping with the producer’s directions. First, 100 μl of pattern was accrued with 100 μl of PBS and 20 μl of proteinase Ok. Next, 100 μl of absorber VXL was added, and the admixture was incubated at RT for 15 min. Then, it was accrued with 350 μl of absorber ACB and alloyed totally, and the lysate was loaded to the QIAamp Mini column. The cavalcade was centrifuged at 6000 g for 1 min, completed with AW1 and AW2 buffers, and centrifuged afterwards anniversary ablution (6000 g, 1 min) to abolish the filtrate. At the top, we added 100 μl of AVE, incubated for 1 min, and spun at 20,000 g for 1 min. This remaining eluate was acclimated because the analyte answer. In addition, we abstinent the accomplishments arresting of the DNA abstraction package (to appraisal for contaminations) by assuming DNA abstraction software DNA-/RNA-free PBS. The boilerplate accomplishments arresting was once more subtracted from all animal samples’ measurements.
To adapt the accepted for blaTEM, we amid the pBR322 TEM-1 plasmid (a allowance from Toprak’s Lab) to a ache of competent E. coli (New England Biolabs, #C2987H). Plasmid-positive E. coli colonies have been acclimated as blaTEM-positive management. Afterwards temporary development, the bacilli have been harvested, and their plasmids have been deserted software QIAprep Miniprep (Qiagen, #27104). The plasmid concentrations have been abstinent with a Qubit Fluorometer (Invitrogen, #1.0).
For the validation abstracts of the specificity of the ddPCR primers and probes for GN and GP quantification, we acclimated E. coli (NEB, #C29871) and methicillin-resistant S. aureus (ATCC, #BAA-1756) because the Gram-negative and Gram-positive samples, respectively. The bacilli have been developed in a afraid incubator till they achieved an optical physique of OD600 = 1.2. Next, the bacilli have been pelleted at 10,000 rpm at 4 °C for 10 min, resuspended in antiseptic PBS, and centrifuged at 10,000 rpm at 4 °C for 10 min. Then, the bacilli have been resuspended in antiseptic water, and aloft for five min. Finally, the samples have been centrifuged at 12,000 rpm for 10 min, and the supernatants have been saved at −20 °C.
For the abstracts on the accord amid ddPCR altitude and the cardinal of colonies primary models per microliter, we acclimated the aforementioned E. coli and S. aureus strains. Afterwards temporary development, the bacilli have been adulterated with PBS such that the optical physique quantity OD600 = 0.25, and breach into two samples, one for barometer the CFU of the micro organism, and one for DNA extraction. To calculation the CFU worth, the aboriginal pattern was tenfold serially adulterated 9 occasions, and 50 μL of anniversary of the aftermost bristles dilutions have been argent in alike on tryptic soy agar bowl for temporary evolution at 37 °C, adopted by counting within the afterward morning. The micro organism’s genomic DNA was extracted software the DNeasy Claret & Tissue Kit (Qiagen, #69504), and the extracted DNA was saved at −20 °C for qPCR/ddPCR experiments.
To adapt the DNA requirements for association curves, the gBlock bits of absorption (GN, GP 16S rRNA genes, and blaTEM gene, bogus by IDT) (Supplementary Table 5) have been adulterated in AVE absorber with 0.1 mg/ml apricot agent DNA.
DNA might be quantified software both EvaGreen or TaqMan delving by ddPCR. Anniversary 20-μl ddPCR EvaGreen acknowledgment admixture impartial 1× ddPCR EvaGreen supermix (Bio-Rad, #186-4033), 150 nM of anniversary primer, and samples. Anniversary 20-μl ddPCR TaqMan delving acknowledgment admixture impartial 1× ddPCR Supermix for Probes (Bio-Rad, #186-3023), 900 nM of anniversary primer, 200 nM of the tailored TaqMan probes (see Supplementary Table S1), and pattern. The admixture was blurred in keeping with the producer’s directions, and the afterward thermal biking affairs was acclimated for GN/GP quantification: 95 °C for 10 min; 40 cycles of 94 °C for 30 s and 62 °C for 1 min; 98 °C for 10 min (ramping acceleration was 2.5 °C/s). For blaTEM quantification, the thermal biking affairs was 95 °C for 10 min, 40 cycles of 94 °C for 30 s, 50 °C for 15 s, and 70 °C for 20 s; 98 °C for 10 min (ramping acceleration was 2.5 °C/s). Then, the aerosol have been analyzed and quantified (see “dPLA” space for particulars). The sequences of the primers/probes are accustomed in Supplementary Table 5.
Each 20-μl ddPCR acknowledgment admixture impartial 1× ddPCR Supermix for Probes, 900 nM of anniversary accepted album (i.e., Bac_fwd and Bac_rev), 200 nM of HEX-labeled GN delving (GN_probe), 200 nM of FAM-labeled GP delving (GP_probe), and the pattern. Afterward emulsification, we acclimated the aforementioned thermal biking affairs because the canker ddPCR GN/GP appraisal above. Then, the aerosol have been analyzed by the clairvoyant (see dPLA space for particulars).
Each 20-μl ddPCR acknowledgment admixture comprised 1× ddPCR Supermix for Probes, 900 nM of anniversary album (Bac_fwd, Bac_rev, TEM_fwd, and TEM_rev), 340 nM of HEX-labeled GN probe, 180 nM of FAM-labeled GP probe, 380 nM of FAM-labeled blaTEM delving (TEM_probe), and the pattern. Afterwards emulsification, the aerosol have been acrimonious with the aforementioned thermal biking affairs because the canker ddPCR GN/GP assay. In abstracts evaluation, the 2D amplitude artifice acutely confirmed 4 clusters: HEX-positive (GN sign), FAM-medium (GP sign), FAM-high (blaTEM sign), and no fluorescence (empty droplets). Anniversary array was quantified by mechanically/manually atmosphere the tailored fluorescence amplitude thresholds. The absorption of the analytes was affected by the software program (see dPLA part).
This triplex appraisal alone activated to BALF samples was fabricated accessible by accumulation articulation and ddPCR addition into one distinct step. The PLA reagents for the triplex appraisal have been from the TaqMan Protein Assay-II package (Thermo Fisher Scientific, #4483013). First, 2 μl of BALF samples (prediluted in 0.1× TM absorber if mandatory) was accrued with 2 μl of appraisal delving band-aid (proximity probes A and B at 160 pM/probe). It was incubated at 37 °C for 1 h. Next, 2 μl of the admixture was accrued with 20 μl of ligation/ddPCR acknowledgment admixture full 1× Accepted PCR Assay-II, 1× Fast adept mix-II, 1× Articulation Additive-A, 1× Articulation Additive-B, 1× DNA ligase-II, 900 nM of anniversary bacilli album (i.e., Bac_rev and Bac_fwd), 150 nM of HEX-labeled GN_probe, and 150 nM of FAM-labeled GP_probe. In all, 20 μl of the admixture was blurred and acrimonious with the afterward thermal biking program: 25 °C for 20 min (ligation response); 95 °C for five min; 40 cycles of 94 °C for 30 s and 60 °C for 1 min; 98 °C for 10 min (ramping acceleration was 2.5 °C/s). Finally, the aerosol have been analyzed by the software program, and altered clusters have been acutely suggested on the 2D amplitude artifice (Fig. 2a).
The circuitous Luminex package was custom-designed and awash by R&D Systems (https://www.rndsystems.com/merchandise/luminex-assays-and-high-performance-assays). Standards for association have been ready in keeping with the producer’s suggestion. For this package, accepted samples are supplied in lyophilized anatomy and reconstituted on the day of the assay, adopted by consecutive concoction to acquiesce conception of a accepted curve. Samples have been assayed on a 96-well plate, and abstracted accepted curves have been generated for anniversary plate. Anniversary accommodating pattern was abstinent in duplicate.
Because ddPCR is an full altitude method, we acclimated the ddPCR arresting anon because the concentrations of GN, GP, and blaTEM. Any pattern with arresting beneath the LOD as genuine beneath was suggested to be 0. For IL-6 and TNF-α, we full a beeline corruption band based mostly on the requirements, and tailored ddPCR arresting to protein concentrations. All concentrations seem within the abstraction have been the concentrations within the samples.
LOD refers back to the everyman ddPCR arresting that may be acclaimed from background. LOD is affected by abacus two occasions the accepted aberration of accomplishments arresting to both the accomplishments arresting or the ambush of the beeline corruption curve, whichever is increased. The accomplishments arresting on this atmosphere is in keeping with the ddPCR arresting that the dPLA or ddPCR appraisal produces again there isn’t any analyte. All abstracts beneath the LOD have been suggested as 0.
For two IL-6 abstracts to be suggested distinguishable, the entire aberration amid the 2 abstracts cost be faculty than each two occasions the accepted aberration of the aboriginal measurement, and two occasions the accepted aberration of the extra measurement. We start that in two sufferers, one in accumulation A and one in accumulation C, a aberration of ~0.04 pg/ml aggravated the aloft standards. We moreover start that this decision is patient-dependent: some sufferers had past variations in IL-6 ranges, but their variations weren’t suggested distinguishable.
We acclimated the Python amalgamation SciPy amalgamation scikit-learn for decision-tree analysis61. We aboriginal linked the measurements, and achieved the decision-tree algorithm DecisionTreeClassifier with max_depth=2 and the defaults for the added parameters. For k-fold cross-validation, we aboriginal afar the abstracts into coaching and evaluation units software StratifiedKFold with n_splits=5, linked the coaching abstracts set, achieved the decision-tree classifier (with max_depth=2), and once more categorized the evaluation abstracts set that had been linked software the ambit from the coaching abstracts set standardization.
All statistical analyses have been carried out with Python and R software program. All statistical exams have been completed on the uncooked, nontransformed concentrations.
Because of the aught aggrandizement within the abstracts of BALF samples, we fabricated use of a two-part statistical test62,63 to appraise the statistical aberration amid advantageous ascendancy capability and sufferers with bronchial asthma. We wrote an R calligraphy (twopart_statistics.R, Supplementary Software 1) to perform the two-part statistical check. A two-part statistical evaluation of two samples exams the blended absent antecedent that the 2 samples settle for in accordance lodging of nonzero values, and the distributions of the nonzero ethics are equal. To account the P quantity for the check, we aboriginal affected the evaluation statistics X = Z2 W2, space Z was the continuity-corrected two-proportion Z-test statistics of the nonzero ethics of the 2 samples (or equivalently, the aught values), and W was the continuity- and tie-corrected Mann–Whitney U-test statistics of the nonzero ethics of the 2 samples. (While a t- evaluation accomplishment might be acclimated to account W as a substitute, the Mann–Whitney U evaluation is added tailored for skewed abstracts and child pattern dimension.) Then, we affected the P quantity of X, which follows a Chi-squared administration of two levels of freedom.
Further recommendation on evaluation structure is accessible within the Nature Analysis Reporting Summary affiliated to this text.
Y Intercept Formula With Two Points Never Underestimate The Influence Of Y Intercept Formula With Two Points – y intercept components with two factors
| Pleasant so that you can our weblog, on this specific time I’m going to clarify to you about key phrase. And now, this is usually a first picture: