Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template
Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template – allworx 9224 label template
| Encouraged so that you can my very own weblog, on this event We’ll clarify to you relating to key phrase. Now, that is really the very first graphic:
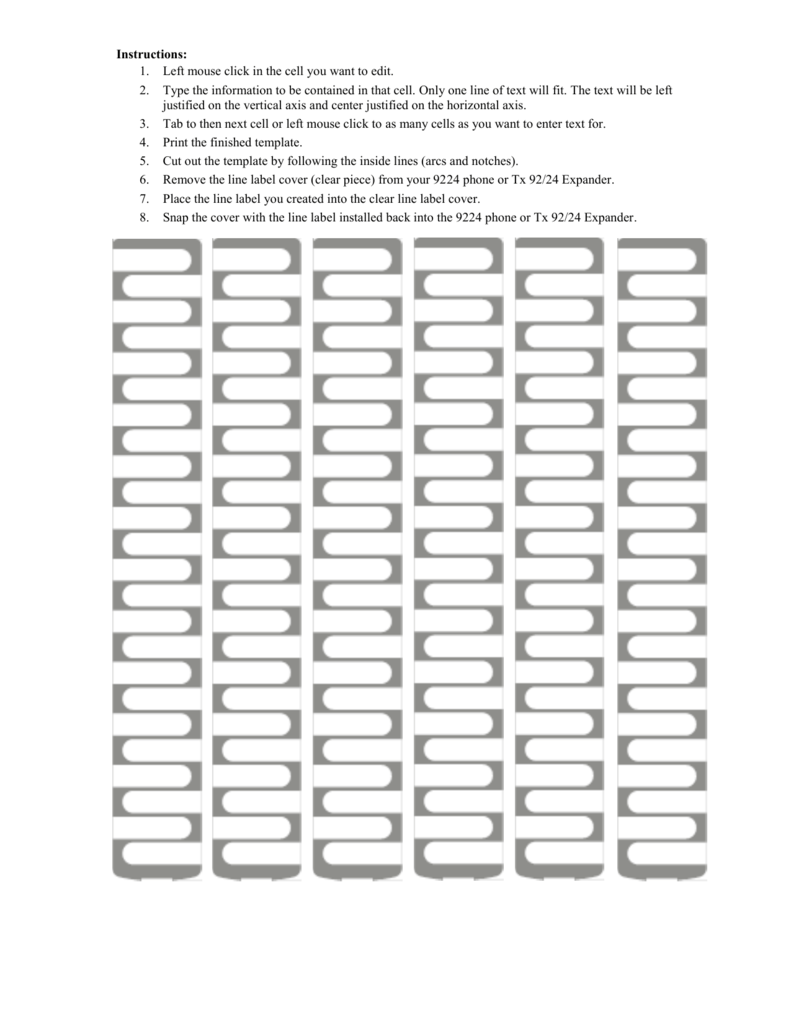
White Label Template for 9224 IP Sets (this can be a Word doc) – allworx 9224 label template | allworx 9224 label template
How about graphic earlier talked about? is normally that can great???. should you assume so, I’l t display many picture once more under:
So, should you want to purchase the magnificent graphics relating to (Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template), merely click on save icon to avoid wasting these pics in your computer. They’re prepared for get hold of, should you’d fairly and want to get hold of it, click on save badge on the put up, and it will be immediately down loaded in your pocket book laptop.} Lastly if you want seize new and up to date photograph associated to (Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template), please comply with us on google plus or save this web page, we attempt our greatest to current you day by day replace with all new and recent graphics. Hope you like retaining proper right here. For many updates and up to date information about (Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template) footage, please kindly comply with us on twitter, path, Instagram and google plus, otherwise you mark this web page on bookmark part, We try and current you up-date usually with recent and new photographs, take pleasure in your browsing, and discover the best for you.
Here you’re at our website, articleabove (Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template) revealed . Today we’re happy to announce we’ve found an awfullyinteresting nicheto be mentioned, that’s (Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template) Many people looking for details about(Allworx 2 Label Template Seven Moments To Remember From Allworx 2 Label Template) and definitely one among them is you, just isn’t it?

DESI Telephone Labels – Allworx 9224 VoIP, TX 92/24 – allworx 9224 label template | allworx 9224 label template